ImageNomer Documentation
Getting Started - Live Web Demo
There is a live web demo running at https://aorliche.github.io/ImageNomer/live/
This demo contains a single Fibromyalgia dataset from OpenNeuro.org, the same as contained in the Docker image.
Once you have loaded ImageNomer, you can check out the Fibromyalgia Dataset Tutorial.
Note: The ImageNomer live demo retains state for the duration of the session.
This includes generated correlation images, which may impact stability if many users access the web demo in a short amount of time.
Getting Started with Docker
Install docker
You need to install docker for this to work. Refer to, e.g., https://www.docker.com/ for instructions on how to install for your operating system.
Download and run the docker image
docker pull ghcr.io/aorliche/image-nomer:latest
docker run -p 8008:8008 ghcr.io/aorliche/image-nomer:latest
We expose port 8008 of the container to be accessible from your web browser.
Note: If you are using Apple Silicon, use the image-nomer-arm64 image instead.
In your browser, navigate to http://localhost:8008/
Explore example data
We have provided an fMRI study of Fibromyalgia from OpenNeuro.org for you to explore.
Unfortunately, due to NIH data policy, we cannot provide access to the Philadelphia Neurodevelopmental Cohort (PNC) dataset or to BSNIP. Access may be obtained for research purposes through the database of Genotypes and Phenotypes (dbGaP). If you do have permission, we would be happy to work with you regarding functions such as, e.g. SNPs, that are not available in the Fibromyalgia dataset.
Adding Your Own Data to Docker Image
You can map a local directory containing your own data into the Docker image. This directory should contain a “demographics.pkl” file as well as an “fc” subdirectory.
We have provided a second example dataset ds004775 from OpenNeuro.org to demonstrate mapping a local data directory into the Docker image.
First, navigate to the “examples” directory of the ImageNomer GitHub repository in your browser.
Download and unzip the included zip file.
To start the Docker image with inclusion of the data you just unzipped, execute the following command:
docker -run -p 8008:8008 \
-v /full/path/to/my/unzipped/cohort:/root/ImageNomer/data/VicariousPunishment \
ghcr.io/aorliche/image-nomer:latest
Navigate to http://localhost:8008/, or, if already there, refresh the page.
The new cohort should be available under the dropdown menu.
See the ImageNomer26FibromyalgiaDataset.ipynb notebook file for an example of how to import data into ImageNomer format when starting with a csv file and BOLD timeseries.
Also see the Punish2FC_ExampleImNomer.ipynb notebook file for an example of a smaller dataset. The “Punish1SPM.ipynb” notebook file in the same directory will show how to use nipype/SPM to create an ImageNomer dataset from 4D fMRI images.
Run By Cloning GitHub Repository
You may also add your own data by cloning the GitHub repository and installing the required Python dependencies.
git clone https://github.com/TulaneMBB/ImageNomer
cd ImageNomer
pip install -r requirements.txt
The repository already contains the Fibromyalgia dataset. To run ImageNomer, execute the following command:
python backend/app.py
Then, navigate to http://localhost:8008
Data is stored in the “ImageNomer/data” directory. Each cohort has its own subdirectory. See the ImageNomer26FibromyalgiaDataset.ipynb notebook file for an example of how to import data into ImageNomer format when starting with a csv file and BOLD timeseries.
Changing the Code
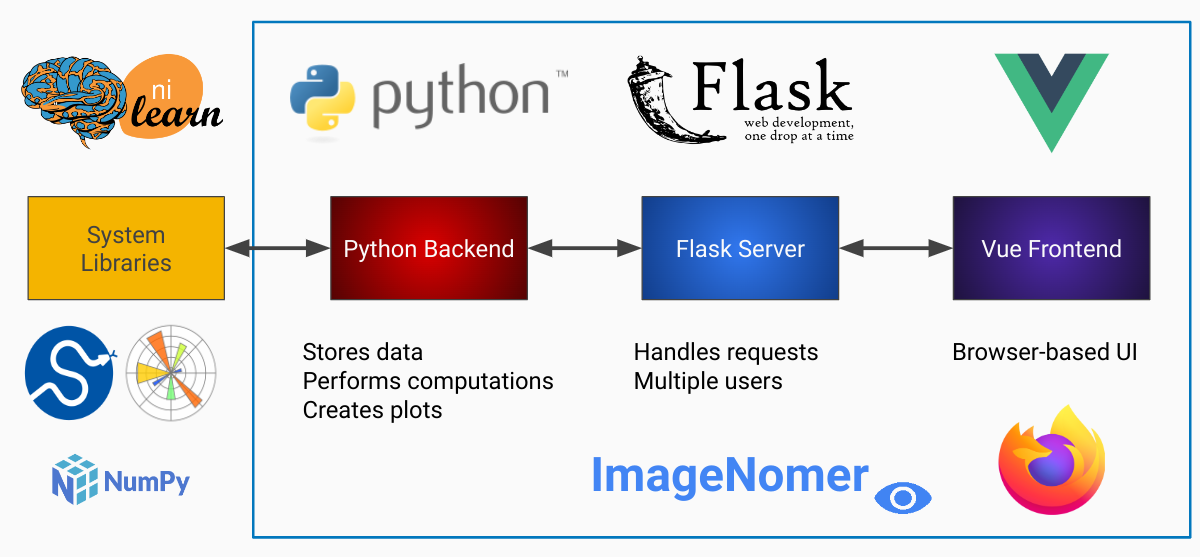
ImageNomer consists of a a Python backend and a Vue javascript frontend. All Python packages are listed in the requirements.txt file. Vue requirements can be installed with npm from the frontend directory.
Please reach out to me with any questions.
Known Bugs
We know that sometimes one or more FC or PC images in the FC/PC view may get stuck on “loading”. This seems to be a problem with the Vue frontend code. We are working on a fix.